Publications
Wang, W., Walmacq, C., Chong, J., Kashlev, M.* & Wang, D.* (2018) Structural basis of transcriptional stalling and bypass of abasic DNA lesion by RNA polymerase II. Proc. Natl. Acad. Sci. USA. (published ahead of print)
Local, A., Huang, H., Albuquerque, C.P., Singh, N., Lee, A.Y., Wang, W., Wang, C., Hsia, J.E., Shiau, A.K., Ge, K., Corbett, K.D., Wang, D., Zhou, H. & Ren, B. (2018) Identification of H3K4me1-associated proteins at mammalian enhancers. Nat. Genet. 50(1):73-82.
Xu, J., Lahiri, I., Wang, W., Wier, A., Cianfrocco, M.A., Chong, J., Hare, A.A., Dervan, P.B., DiMaio, F., Leschziner, A.E.* & Wang, D.* (2017) Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature. 551:653-657.
Featured in UCSD blog post:
http://ucsdhealthsciences.tumblr.com/post/167774555205/frozen-in-action-how-cells-repair-dna-as-its
Wang, W., Xu, L., Hu, L., Chong, J., He, C. & Wang, D.* (2017) Epigenetic DNA modification N6-methyladenine causes site-specific RNA polymerase II transcriptional pausing. J. Am. Chem. Soc.139(41):14436-14442.
Xu, L., Wang, W., Wu, J., Shin, J.H., Wang, P., Unarta, I.C., Chong, J., Wang, Y.* & Wang, D.* (2017) Mechanism of DNA alkylation-induced transcriptional stalling, lesion bypass, and mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 114(34):E7082-E7091.
Cui, M., Feng, H., Guo, D., Wang, D. & Zheng, B.* (2017) A droplet-based microfluidic platform for measuring both rapid and slow kinetics. Sensor. Actuat. B-Chem. 253:731-737.
Shin, J.H., Xu, L. & Wang, D.* (2017) Mechanism of transcription-coupled DNA modification recognition. Cell Biosci. 7:9.
Xu, L., Wang, W., Gotte, D., Yang, F., Hare, A.A., Welch, T.R., Li, B.C., Shin, J.H., Chong, J., Strathern, J.N.*, Dervan, P.B.* & Wang, D.* (2016) RNA polymerase II senses obstruction in the DNA minor groove via a conserved sensor motif. Proc. Natl. Acad. Sci. U.S.A. 113(44):12426-12431.
Yamamoto, K., Wang, J., Sprinzen, L., Xu, J., Haddock, C.J., Li, C., Lee, B.J., Loredan, D.G., Jiang, W., Vindigni, A., Wang, D., Rabadan, R. & Zha, S.* (2016) Kinase-dead ATM protein is highly oncogenic and can be preferentially targeted by Topo-isomerase I inhibitor. eLife. pii:e14709
Hwang, C.S., Xu, L., Wang, W., Ulrich, S., Zhang, L., Chong, J., Shin, J.H., Huang, X., Kool, E.T., McKenna, C.E.* & Wang, D.* (2016) Functional interplay between NTP leaving group and base pair recognition during RNA polymerase II nucleotide incorporation revealed by methylene substitution. Nucleic Acids Res. 44(8):3820-8.
Shin, J.H., Xu, L. & Wang, D.* (2016) RNA polymerase II acts as a selective sensor for DNA lesions and endogenous DNA modifications. Transcription. 7(3):57-62. (Invited review)
Zhang, L., Pardo-Avila, F., Unarta, I.C., Cheung, P.P., Wang, G., Wang, D. & Huang, X.* (2016) Elucidation of the dynamics of transcription elongation for RNA polymerase II using kinetic network models. Acc. Chem. Res. 49(4):687-94.
Da, L.T., Pardo-Avila, F., Xu, L., Silva, D.A., Zhang, L., Gao, X., Wang, D.* & Huang, X.* (2016) Bridge helix bending promotes RNA polymerase II backtracking through a critical and conserved threonine residue. Nat. Commun. 7:11244.
Xu, L., Wang, W., Chong, J., Shin, J.H., Xu, J. & Wang, D.* (2015) RNA polymerase II transcriptional fidelity control and its functional interplay with DNA modifications. Crit. Rev. Biochem. Mol. Biol. 50(6):503-19.
Zhang, L., Silva, D.A., Pardo-Avila, F., Wang, D. & Huang, X.* (2015) Structural model of RNA Polymerase II elongation complex with complete transcription bubble reveals NTP entry routes. PLoS Comput. Biol. 11(7):e1004354.
Wang, L., Zhou, Y., Xu, L., Xiao, R., Lu, X., Chen, L., Chong, J., Li, H., He, C., Fu, X-D.* & Wang, D.* (2015) Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature. 523(7562):621-25.
Xu, L., Wang, W., Zhang, L., Chong, J., Huang, X. & Wang, D.* (2015) Impacts of template backbone heterogeneity on RNA polymerase II transcription. Nucleic Acids Res. 43(4):2232-41.
Walmacq, C., Wang, L., Chong, J., Scibelli, K., Lubkowska, L., Gnatt, A., Brooks, P.J., Wang, D.* & Kashlev, M.* (2015) Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions. Proc. Natl. Acad. Sci. U.S.A. 112(5):E410-9.
Wang, L., Limbo, O., Fei, J., Chen, L., Kim, B., Luo, J., Chong, J., Conaway, R.C., Conaway, J.W., Ranish, J.A., Kadonaga, J.T., Rusell, P. & Wang, D.* (2014) Regulation of the Rhp26ERCC6/CSB chromatin remodeler by a novel conserved leucine latch motif. Proc. Natl. Acad. Sci. U.S.A. 111(52):18566-71.
Xu, L., Chen, Y.-C., Chong, J., Fin, A., McCoy, L.S., Xu, J., Zhang, C. & Wang, D.* (2014) Pyrene-based quantitative detection of the 5-formylcytosine loci symmetry in the CpG duplex content during TET-dependent demethylation. Angew. Chem. Int. Ed. Engl. 53(42):11223-7.
Xu, L., Zhang, L., Chong, J., Xu, J., Huang, X. & Wang, D.* (2014) Strand-specific (asymmetric) contribution of phosphodiester linkages on RNA polymerase II transcriptional efficiency and fidelity. Proc. Natl. Acad. Sci. U.S.A. 111(32):E3269-76.
Eun, C.*, Ortiz-Sánchez, J.M., Da, L., Wang, D.* & McCammon, J.A. (2014) Molecular dynamics simulation study of conformational changes of transcription factor TFIIS during RNA polymerase II transcriptional arrest and reactivation. PLoS One. 9(5):e97975.
Xu, L., Butler, K.V., Chong, J., Wengel, J., Kool, E.T. & Wang, D.* (2014) Dissecting the chemical interactions and substrate structural signatures governing RNA polymerase II trigger loop closure by synthetic nucleic acid analogues. Nucleic Acids Res. 42(9):5863-70.
Xu, L., Da, L., Plouffe, S.W., Chong, J., Kool, E.* & Wang, D.* (2014) Molecular basis of transcriptional fidelity and DNA lesion-induced transcriptional mutagensis. DNA Repair (Amst). 19:71-83.
Silva, D.A., Weiss, D.R., Pardo Avila, F., Da, L-T., Levitt, M., Wang, D.* & Huang, X.* (2014) Millisecond dynamics of RNA polymerase II translocation at atomic resolution. Proc. Natl. Acad. Sci. U.S.A. 111(21):7665-7670.
Xu, L., Chen, Y-C., Nakajima, S., Chong, J., Wang, L., Lan, L., Zhang, C.* & Wang, D.* (2014) A chemical probe targets DNA 5-formylcytosine sites and inhibits TDG excision, polymerases bypass, and gene expression. Chem. Sci. 5(2):567-574.
Xu, L., Plouffe, S.W., Chong, J., Wengel, J. & Wang, D.* (2013) A chemical perspective on transcriptional fidelity: dominant contributions of sugar integrity revealed by unlocked nucleic acids. Angew. Chem. Int. Ed. Engl. 52(47):12341-5.
Kellinger, M.W., Park, G.Y., Chong, J., Lippard, S.J.* & Wang, D.* (2013) Effect of a monofunctional phenanthriplatin-DNA adduct on RNA polymerase II transcriptional fidelity and translesion synthesis. J. Am. Chem. Soc. 135(35):13054-61.
Zhang, S. & Wang, D.* (2013) Understanding the Molecular Basis of RNA Polymerase II Transcription. Isr. J. Chem. 53(6-7).
Da, L-T., Pardo Avila, F., Wang, D. & Huang, X. (2013) A two-state model for the dynamics of the pyrophosphate ion release in bacterial RNA polymerase. PLoS Comput. Biol. 9(4):e1003020.
Kellinger, M.W., Song, C., Chong, J., Lu, X., He, C. & Wang, D.* (2012) 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription.
Nat. Struct. Mol. Biol. 19(8):831-3.
Commented in:
Huang Y., Rao A. (2012) New functions for DNA modifications by TET-JBP. Nat. Struct. Mol. Biol. 19(11):1061-4.
Highlighted in:
UC San Diego Health Sciences Blog
Kellinger, M.W., Ultrich, S., Chong, J., Kool, E.T.* & Wang, D.* (2012) Dissecting chemical interactions governing RNA polymerase II transcriptional fidelity. J. Am. Chem. Soc. 134(19): 8231-40.
Reported in reviews:
Svetlov, V. & Nudler, E. (2013) Basic mechanism of transcription by RNA polymerase II. Biochim. Biophys. Acta. 1829(1):20–28.
Kaplan, C.D. (2013) Basic mechanisms of RNA polymerase II activity and alteration of gene expression in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1829(1):39-54.
Da, L-T., Wang, D.* & Huang, X.* (2012) Dynamics of pyrophosphate ion release and its coupled trigger loop motion from closed to open state in RNA polymerase II. J. Am. Chem. Soc. 134(4): 2399-406.
Reported in reviews:
Svetlov, V. & Nudler, E. (2013) Basic mechanism of transcription by RNA polymerase II. Biochim. Biophys. Acta.1829(1):20–28.
Kaplan, C.D. (2013) Basic mechanisms of RNA polymerase II activity and alteration of gene expression in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1829(1):39-54.
Huang, X., Wang, D., Weiss, D.R., Bushnell, D.A., Kornberg, R.D. & Levitt, M. (2010) RNA polymerase II trigger loop residues stabilize and position the incoming nucleotide triphosphate in transcription. Proc. Natl. Acad. Sci. U.S.A. 107(36):15745-50.
Reported in:
Faculty of 1000 Biology as recommended F1000 Factor 6.0
Wang, D.*, Zhu, G., Huang, X. & Lippard, S.J.* (2010) X-ray structure and mechanism of RNA polymerase II stalled at an antineoplastic monofunctional platinum-DNA adduct. Proc. Natl. Acad. Sci. U.S.A. 107(21): 9584-9589.
Reported in:
Faculty of 1000 Biology as recommended F1000 Factor 8.0
Liu, X., Bushnell, D.A., Wang, D., Calero, G. & Kornberg, R.D. (2010) Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 327(5962):
206-9.
Wang, D., Bushnell, D.A., Westover, K.D., Huang, X., Levitt, M. & Kornberg, R.D. (2009) Structural basis of transcription: backtracked RNA polymerase II at 3.4 Å resolution. Science. 324(5931): 1203-6.
Reported in:
Faculty of 1000 Biology as Must Read F1000 Factor 10.0
ALSNews and SLAC today
Wang, D., Bushnell, D.A., Westover, K.D., Kaplan, C.D. & Kornberg, R.D. (2006) Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 127(5): 941-54. (Cover)
Reported in:
SSRL Science Highlight & Light Source Science Highlight
Danford, A.J., Wang, D., Wang, Q., Tullius, T.D. & Lippard, S.J. (2005) Platinum anticancer drug damage enforces a particular rotational setting of DNA in nucleosomes. Proc. Natl. Acad. Sci. U.S.A. 102(35): 12311-6.
Takagi, Y., Masuda, C.A., Chang, W.H., Komori, H., Wang, D., Hunter, T., Joazeiro, C.A. & Kornberg, R.D. (2005) Ubiquitin ligase activity of TFIIH and the transcriptional response to DNA damage. Mol. Cell. 18(2): 237-243.
Reported in:
Faculty of 1000 Biology as Must Read F1000 Factor 8.0
Wang, D. & Lippard, S.J. (2005) Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 4(4): 307-20. (Review)
Wang, D. & Lippard, S.J. (2004) Cisplatin-induced post-translational modification of histones H3 and H4. J. Biol. Chem. 279(20): 20622-5.
Wang, D., Hara, R., Singh, G., Sancar, A. & Lippard, S.J. (2003) Nucleotide excision repair from site-specifically platinum-modified nucleosomes. Biochemistry. 42(22): 6747-53.
Lee, K.B., Wang, D., Lippard, S.J. & Sharp, P.A. (2002) Transcription-coupled and DNA damage-dependent ubiquitination of RNA polymerase II in vitro. Proc. Natl. Acad. Sci. U.S.A. 99(7): 4239-44.
Guan, R. J., Wang, M., Wang, D. & Wang, D.C. (2001) A new insect neurotoxin AngP(1) with analgesic effect from the scorpion Buthus martensii Karsch: purification and characterization. J. Pept. Res. 58(1): 27-35. (Now known as Chem. Biol. Drug Des.)
Highlights
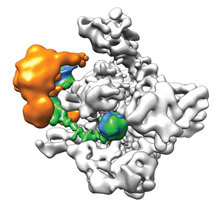
Wang group solved the CryoEM structure of Pol II-Rad 26 complex and elucidated the structural basis of initiation of eukaryotic transcription-coupled DNA repair.
Learn more
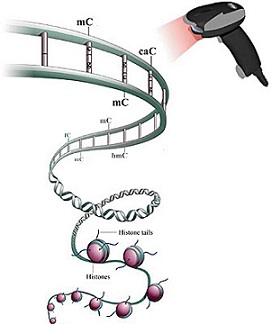
The Wang group solved the crystal structure of Pol II in complex with 5-carboxylcytosine (5caC) and revealed how Poll II recognizes subtle changes in DNA sequence and structure.
Learn more

Using an acyclic synthetic RNA analogue, the Wang group demonstrated that the contribution of the intact nucleotide sugar bakbone to the enzymatic efficiency and transcriptional fidelity of RNA polymerase II is significantly stronger than that of the other functional groups of the nucleotide, including the nucleobase and ribose 2'-OH group.
Learn more
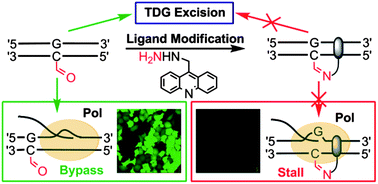
The Wang group designed and constructed a chemical probe, consisting of an aldehyde-targeting group and an intercalation group, that can selectively react with 5-formylcytosine (5fC) and subsequently inhibit base excision by thymine DNA glycosylase (TDG) and cause significant pausing for both DNA and RNA polymerase elongation. Further investigation using a GFP reporter system in living cells revealed that the covalent modification in 5fC sites at 5'-UTR of the GFP gene greatly inhibited the GFP expression level.
Learn more
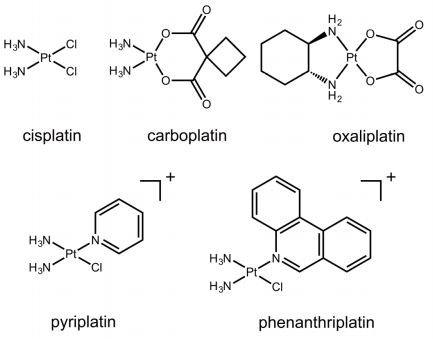
The Wang group present the first systematic mechanistic investigation that addresses how a site-specific phenanthriplatin-DNA d(G) monofunctional adduct affects the Pol II elongation and transcriptional fidelity checkpoint steps, which is an important step toward elucidating the mechanism of action of phenanthriplatin.
Learn more
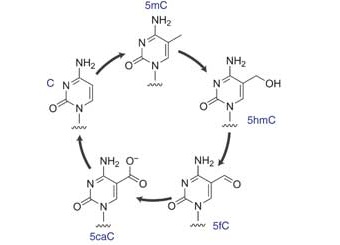
The Wang group reported a systematic study of the effects of five different forms of cytosine in DNA on mammalian and yeast RNA polymerase II transcription.
Learn More
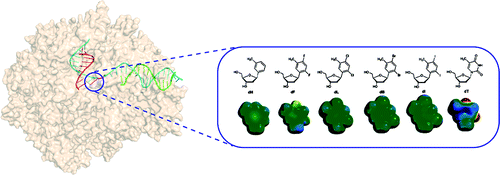
Using a series of "hydrogen bond deficient" nucleoside analogues, the Wang group provided the first systematic evaluation of electrostatic and steric effects in controlling RNA Polymerase II transcriptional fidelity.
Learn More
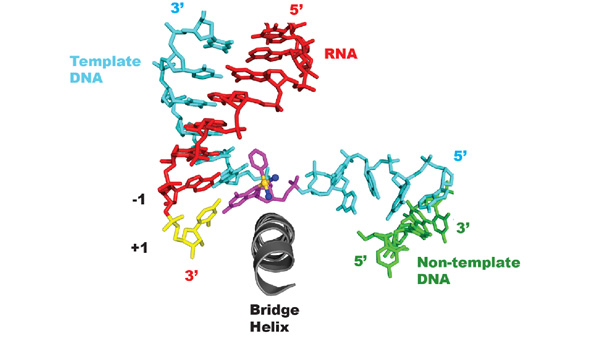
The Wang group reported the structure of a transcribing RNA polymerase II (Pol II) complex stalled at a site-specific monofunctional pyriplatin-DNA adduct in the active site. The results reveal a molecular mechanism of Pol II transcription inhibition and drug action that is dramatically different from transcription inhibition by cisplatin and UV-induced 1,2-intrastrand cross-links. Our findings provide insight into structure-activity relationships that may apply to the entire family of monofunctional DNA-damaging agents and pave the way for rational improvement of monofunctional platinum anticancer drugs.
Learn more






